
A selected region of a digitized H&E-stained slide [Image courtesy of PathAI]
In a recent conversation with Dr. Andy Beck, co-founder and CEO of PathAI, we had the opportunity to discuss PathExplore, an AI-driven platform that aims to transform the way tumor microenvironment (TME) analysis is conducted. Traditional methods such as manual pathology, multiplex immunofluorescence and single-cell omics often face limitations, including high costs or tissue consumption. PathExplore addresses these challenges by using AI to analyze digitized H&E slides, which are widely available and don’t necessitate advanced assays or equipment.
In the following interview, Dr. Beck highlights the advantages of PathExplore, such as its ability to bridge the gap between H&E and high-resolution modalities such as multiplex and single-cell RNA sequencing and its potential for rapidly scaling biomarkers. Additionally, the platform employs human interpretable features (HIFs) to characterize the spatial biology of the TME, enabling correlations with clinical trial outcomes, multi-omics and real-world evidence.
How does PathExplore’s AI-driven approach to analyzing the tumor microenvironment (TME) differ from traditional methods?

Dr. Andy Beck
Beck: There are a few approaches to analyzing the TME, spanning from traditional pathology (without the use of AI) to newer and more specialized approaches such as multiplex immunofluorescence and single-cell omics.
Using traditional, manual pathology to analyze the TME can be quite limited. In most cases, pathologists and researchers are using H&E or IHC-stained slides to look at a specific set of TME features, often within a specific region of interest on a slide, which is fine for certain well-defined hypotheses and investigations, but it’s hard to scale and exhaustively analyze the TME.
It takes some of our own pathologists in our network over 20 hours to manually annotate one-quarter of a whole-slide image, so imagine the impracticality and tediousness to do that not only for an entire image but then for the hundreds or thousands of slides that pharma has in their repertoire of clinical trial samples used for R&D.
What advantages does PathExplore’s AI-driven approach provide compared to manual pathology?
We have very high-resolution methods such as multiplex IF and single-cell omics. mIF panels can be 100+ plex, so you get a massive amount of data, but the technology for mIF is often specialized and expensive. Single-cell omics gives you the molecular profile at a cell level, but you don’t have that connection to the histopathological disease manifestation. On top of that, this process is very time intensive, and it consumes the tissue, i.e., the tissue cannot be reused.
Unlike mplex IF and single-cell and spatial transcriptomics, H&E is already ubiquitous, it doesn’t require more advanced assays and equipment, and it doesn’t destroy the tissue. All we need is to digitize the H&E slides that pharma already has! And then we run the PathExplore algorithm on those images.
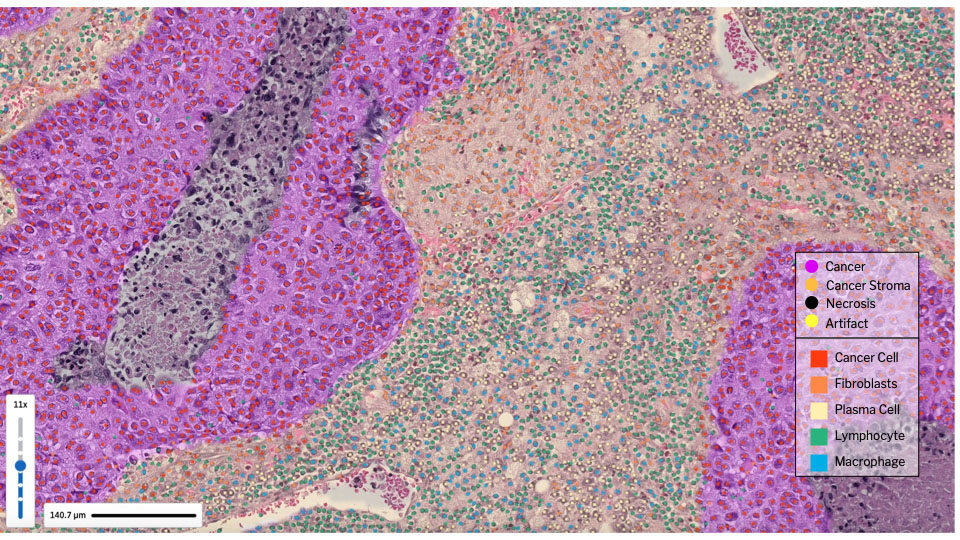
AI-generated tissue heatmaps and cell identification of the tumor microenvironment, overlaid on a digitized image of an H&E slide. [Image courtesy of PathAI]
So, what are the benefits here? First, AI helps to bridge the gap between H&E and high-res modalities such as mplex and single cell RNA seq. AI unlocks the resolution of single cell, spatial biology already in H&E slides for pharma to then use for drug development without a large investment in new expertise, skills, and infrastructure. We think this can enable more open exploration that is less limited to known hypotheses without needing major large new investments in R&D. And number two, if pharma identifies an actionable biomarker, since this runs off of H&E, these biomarkers can be scaled and made accessible fairly quickly and easily compared to the other modalities.
Can you explain the role of human interpretable features (HIFs) in PathExplore and how they enable a more detailed understanding of the TME?
Beck: It’s not enough to just identify the cells and tissue regions in the TME. Research is showing that it is the spatial interactions across these cells and within specific areas of the tissue that matter. HIFs enable us to characterize the spatial biology of the TME, and then correlate these features to outcomes or other multi-omic data such as genetics to strengthen our understanding of the histopathological drivers of response, resistance and drug mechanism of action.
For example, the research below points to the potential significance of stromally-located tumor-infiltrating lymphocytes on patient survival. Location matters — it’s not enough to just measure the number of lymphocytes.
This then begs the question around how many other spatial biomarkers like TILs could exist in the TME? We don’t know — and that’s where HIFs come in. HIFs are, at the moment, over 600 unique metrics that quantify different combinations of area, counts, densities and ratios between cell types across each tissue region. We can measure the proportion of lymphocytes to macrophages in the stroma. We can measure the number of one cell type within a certain proximity to another cell type. And when we run these metrics through combinations of cell and tissue regions — counts of macrophages in stroma, counts of lymphocytes in stroma, counts of macrophages in cancer, counts of lymphocytes in cancer, etc. Do this again for density. Do this again, but now compare the ratio of macrophages to lymphocytes in stroma, and again in cancer. You can see how the number of spatial relationships to quantify is large — and so far, we have standardized 600 of these metrics into our PathExplore panel of HIFs.

H&E-stained whole slide image of non-small cell lung cancer (left). AI-generated cell and tissue classification heatmaps and overlays on the same whole-slide image (right). [Image courtesy of PathAI]
How is PathExplore’s single-cell resolution of novel spatial signatures predictive of outcomes in cancer patients?
Beck: To recap — the first step is to have AI identify each cell and tissue region. Then we quantify all the various spatial interactions into 600 standardized and structured metrics called HIFs. We essentially have translated an unstructured image into structured outputs/data that can be used in analytical software to correlate HIFs to clinical trial outcomes, multi-omics, and real-world evidence. One application that we see is the use of graph-neural networks to analyze dense high-resolution heatmaps of tissue regions and cell type and location across hundreds or thousands of patient samples and identify patterns in the spatial orientation of cells in the TME. Then we can correlate these spatial, GNN-generated clusters to outcomes to see if they are, in fact, predictive of a specific outcome. There could be a world where we identify a biomarker that isn’t a single biomarker like many are today, e.g., PD-L1, but a cluster of features that are common across multiple patients and predictive of response to a certain therapy.
Here is a link to our article published in Nature Communications where HIFs predicted molecular markers such as HRD. This article is one of the first instances where we coined ‘human interpretable features’ from AI-powered TME analysis.
How does the scalability of the PathExplore platform improve the efficiency of drug development in comparison to current technologies?
Beck: One of the benefits of having standardized HIFs is that researchers can identify a new finding in one indication or program, and then replicate that analysis very quickly across other indications and programs to see if that same biomarker or mechanism of action can be scaled. The other benefit is you can test multiple hypotheses, run many investigations rapidly and repeatedly using the same data. You don’t have to keep running PathExplore for each new investigation. One data output can power countless analyses without ever having to go back and access the tissue or run the algorithm.
How can PathExplore’s standardized and reproducible measures of counts, densities, areas, and spatial relationships across cell types?
Beck: The practical benefit is giving researchers a common language, a common, comparable set of metrics to analyze the TME. If we are all analyzing the TME in different ways, it makes it challenging to compare apples to oranges. HIFs allow more apples-to-apples comparisons of TME investigations, and can allow researchers to build upon past work as well.
The scientific benefit is explainability and interpretability. When we have identified certain HIFs, or signatures, of HIFs that are highly correlated with a certain outcome, we can look at those HIFs and further dig into the biological meaning into explaining why that connection is happening. HIFs eliminate some of the ‘black box’ that exists in AI analyses of biology because all these features are interpretable.
With PathExplore currently available for multiple cancer types, what are your plans for expanding its applications to other cancers and diseases?
Beck: Within oncology, we are planning to expand PathExplore into five additional indications by the end of the year. Outside of oncology, we will be launching a similar HIF panel in NASH and in ulcerative colitis over the next couple of months, which is particularly exciting given the unmet needs in therapeutic development in these two diseases.
In the long term, we believe that PathExplore’s AI-driven approach and HIFs can be applied to any disease that has a tissue component, whether it be infectious diseases, autoimmune diseases, or even rare diseases. The potential applications are vast, and we are excited to continue expanding the use of PathExplore in new areas of research and drug development.