
Image courtesy of GSK.
Pharmaceutical and biotechnology organizations have made significant progress enabling accelerated drug discovery with traditional laboratory and computing methods. However, they have only just begun to harness the potential of artificial intelligence (AI) to develop more effective therapeutics faster.
This article looks at recent work from researchers at GlaxoSmithKline (GSK) with Cerebras Systems as an example of how the right AI computing platform can enable new research and new methods and potentially transform the future of drug discovery.
Inside the GSK and Cerebras partnership
GSK and Cerebras Systems have a partnership that uses AI computing from Cerebras to accelerate pharmacological and life sciences research at GSK. As a recent example of the power of this partnership, GSK researchers worked with Cerebras Systems to develop a new AI model of cell-type-specific gene regulation and function based on both DNA sequence and epigenetic data.
The new AI model from GSK is named Epigenomic BERT or “EBERT,” and is based on a well-known natural language processing (NLP) model named BERT (Bidirectional Encoder Representations from Transformers), originally developed by Google researchers in 2018. While it may seem counterintuitive for a model built for human language and written text to represent gene regulation and cellular function, in this case, the genetic and other biochemical sequence data that the team employed can be thought of as dialects of the “language” of biology. Instead of predicting the answer to a question online, for example, the GSK EBERT model can predict transcription factor binding sites in cells to inform the potential development of genetically validated therapeutics for human health problems.
Deep learning models like this have great potential to inform and accelerate drug development to improve health outcomes. However, they are computationally intensive and not easy to work with using traditional computer clusters. It can often take researchers weeks or longer to train a single model. In addition, many such model training experiments need to be run iteratively to home in on the combination of model parameters that best represent underlying phenomena. Whether you are a researcher testing many hypotheses or an application developer building a model for a business production case, this is too long.
The EBERT model by GSK is such a model. It employs a unique and large dataset combining DNA data – similar to the previous DNABERT (or “DBERT”) model – and added epigenetic data from the IDEAS36 database. The primary model trained on those datasets was then additionally trained (or “fine-tuned”) on transcription factor binding datasets developed for the ENCODE-DREAM project to evaluate the usefulness of their new EBERT model compared to earlier DBERT for a set of standard benchmark modeling tasks.
The engineering and computing requirements for all those training runs add up. As such, this is where GSK’s partnership with AI computer systems company Cerebras Systems comes in. Cerebras has built a new processor and new class of computer system to accelerate deep learning by orders of magnitude beyond legacy systems composed of graphics processors. In Cerebras, GSK saw an opportunity to accelerate their use of AI and time to insight and employed a Cerebras CS-1 computer system for their work on EBERT.
GSK’s work on EBERT was enabled by and run on a Cerebras CS-1 system, powered by the Cerebras Wafer-Scale Engine (WSE) – the world’s largest and most powerful processor for deep learning. The first generation WSE within the CS-1 has a massive array of 400,000 programmable AI-optimized cores, 40GB of fast on-chip memory, and a software-configurable high-bandwidth on-wafer core-to-core interconnect – this is a machine built from the ground up for AI work like EBERT.

Above: The Cerebras WSE-1, with an Nvidia graphics processing unit (GPU) for scale.
Conventional AI systems require a large number of graphics processing units (GPUs) to deliver equivalent performance. Trying to split a workload across a dozen or more GPUs also comes with drawbacks.
Scaling out AI models requires large amounts of compute capability, high communication bandwidth between processing elements, and large memory pools. The memory and communication bottlenecks intrinsic to large GPU clusters can make them challenging to program and less efficient at scale, posing inherent challenges for researchers to quickly test many new ideas. The Cerebras CS-1 system allowed GSK to avoid this problem.
The research team trained EBERT in 2.5 days aboard the Cerebras CS-1, a process they estimate would have taken 24 days on a 16-node GPU cluster. Continuing their work with Cerebras in the future, there is an opportunity to go even faster to uncover potentially higher accuracy solutions or other new models. The recently announced second-generation Cerebras System named the CS-2 is powered by a new 850,000 core Wafer-Scale Engine (WSE-2) that delivers more than double the AI compute performance of the CS-1 used for GSK’s current work. In continued partnership with Cerebras, GSK will use the CS-2 to further accelerate EBERT training and related research, bringing new models, new datasets, and new AI to bear on some of the field’s most pressing questions.
Because programming the CS-1 system took less time than programming an equivalent GPU cluster, the research team could spend more time fine-tuning the EBERT model for optimal performance. The tuned version of the model, EBERT+, showed substantial improvements over both EBERT and DBERT.

Transcription factor: cell type
While the degree of improvement from DBERT to EBERT to EBERT+ varies depending on the transcription factor and cell type, all data points are in the same direction. Training on an additional data set and fine-tuning the new EBERT model both helped improve the model’s predictive accuracy.
The table below shows how EBERT+ fared on the ENCODE-DREAM benchmark compared to other AI models. While it did not win the benchmark outright, EBERT+ took third place overall and notably won first place in 4 of the 13 subtests when evaluated against models built specifically for those tasks.
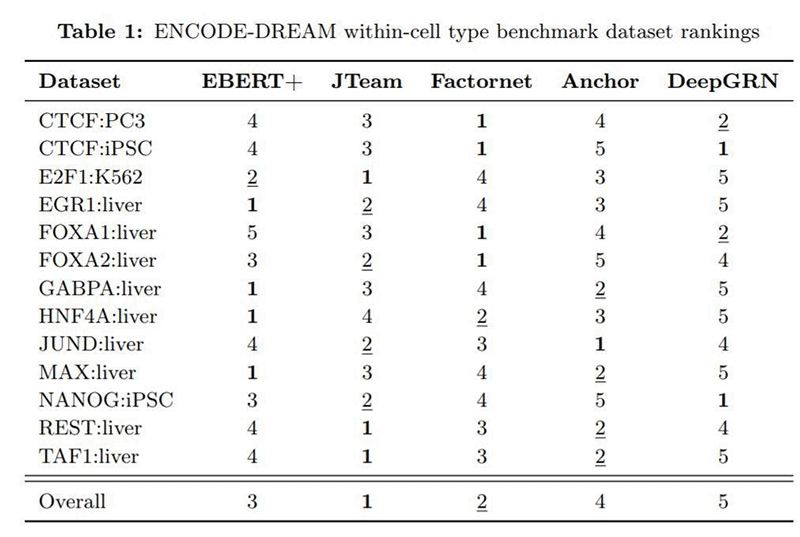
Using conventional hardware, it would have taken days or weeks to make certain EBERT+ could scale properly across multiple GPUs. The Cerebras CS-1 system allowed the team to spend this time fine-tuning the model for higher accuracy instead.
Perhaps just as importantly, GSK was able to test the hypothesis that adding epigenomic data improves the model and that EBERT+ produces better results. As the researchers mention in their arxiv paper, “training these complex models has previously been a computationally intractable problem.” And moreover that, “the training speedup afforded by the Cerebras system enabled us to explore architecture variations, tokenization schemes and hyperparameter settings in a way that would have been prohibitively time and resource-intensive on a typical GPU cluster.” Here, the CS-1 enabled faster computation and unlocked new experimental abilities and a new modeling capability for genetic medicine.
Contemplating AI’s potential in drug discovery
One of the ways that AI could transform medical research and treatment development is by enabling more accurate digital models of biological processes; such digital models of real-world processes are sometimes referred to as “digital twins.” With an accurate digital model of a candidate therapeutic and its biological response, researchers can more easily and quickly explore the universe of treatments and outcomes. Expanding the size of the training data and incorporating information from multiple data sets can help researchers test predicted outcomes with more confidence that their simulated results will match reality. As with the EBERT work described above, enabling this work has significant computational requirements – requirements that can be uniquely addressed by purpose-built AI systems like Cerebras’.
Other discoveries in the field, unrelated to work done by GSK and Cerebras, illustrate the approach’s long-term potential. For example, in 2020, AI identified the drug halicin as a new antibiotic without assistance from human researchers.
In 2021, the German biotech company Evotec announced clinical trials of a new anticancer drug discovered by applying AI principles to drug discovery. AI models like AlphaFold2 and RoseTTAFold can predict a protein’s 3-dimensional folded shape from its base state with far better accuracy than any previous approach.
These breakthroughs and achievements have driven a veritable gold rush into the industry. Firms like Amgen and Sanofi have announced multi-billion dollar deals with machine learning specialist companies like Generate and Exscientia. Both deals are intended to accelerate drug development and discover which molecular compounds are the most promising for treating various diseases.
But the companies streaming into this space face significant bottlenecks if they limit themselves to conventional GPU deployments. The enormous data sets and models expected to power discoveries strain GPU deployments and the programmers who develop them. This problem will only grow in the future as training sets expand, and models become more complex.
AI accelerator systems like the Cerebras CS-1 and its successor, the CS-2, have a vital part to play in this discovery process. One of the strengths of Cerebras’ approach is that it frees researchers to spend more time on cutting-edge science and less time wrestling the difficulty of scaling workloads across a GPU cluster.
These breakthroughs highlight AI’s potential to transform medicine and medical research. A properly trained, sufficiently detailed AI model can simulate the molecular structure of potential medicines far more quickly and effectively than any lab, reducing development time and helping scientists make sure they have identified the most effective compound for treating a given disorder or condition. The long-term implications of these breakthroughs for medicine and pharmacology are profound – and profoundly necessary, given the reality of rising antibiotic resistance and the dearth of options for addressing this problem.

Andy Hock
Andy Hock is vice president and head of product at Cerebras Systems. He and his team lead hardware, software, and machine learning research product requirements and strategy, working with both engineering and customers to build systems that radically accelerate AI for research and enterprise applications. Previously, Andy was the data product lead for the Terra Bella project at Google (née Skybox Imaging), building AI-powered data products for enterprise from satellite imagery, and senior director of Advanced Technologies at Skybox Imaging. Earlier, Andy was a senior technical program manager and senior scientist with Arete Associates. He holds a Ph.D. in geophysics and space physics from UCLA.